Scirpophaga incertulas
the yellow stem borer or rice yellow stem borer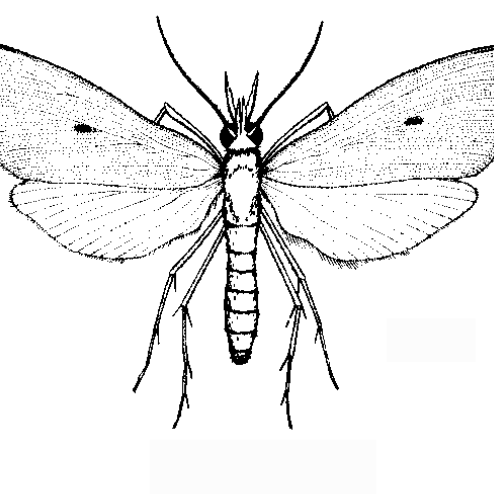
Scirpophaga incertulas, the yellow stem borer or rice yellow stem borer, is a species of moth of the family Crambidae. It was described by Francis Walker in 1863.
Host Genome
| Genome ID | Level | BUSCO Assessment |
|---|---|---|
| - | Chromosome |
C:95.4%[S:88.8%,D:6.6%],F:1.5%,M:3.1%,n:1367
|
Related Symbionts
22 recordsSymbiont records associated with Scirpophaga incertulas
| Classification | Function | Function Tags | Reference | |
|---|---|---|---|---|
|
Microbacterium arborescens DSM 2None754
Actinomycetota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Microbacterium arborescens DSM 2None754
Actinomycetota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Microbacterium oleivorans CCGE2277
Actinomycetota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Microbacteriaceae bacterium BMC-3
Actinomycetota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Microbacteriaceae bacterium BMC-3
Actinomycetota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Microbacteriaceae bacterium HLB-6
Actinomycetota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Microbacterium arborescens JB8_2B
Actinomycetota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Pantoea agglomerans WAB1927
Pseudomonadota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Enterobacter species DHM1T
Pseudomonadota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Klebsiella pneumoniae L-13
Pseudomonadota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Klebsiella pneumoniae L-13
Pseudomonadota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Bacillus subtilis C1YNoneNone1
Bacillota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Bacillus species BB2_1A
Bacillota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Pantoea species NCCP116
Pseudomonadota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Serratia marcescens PS1
Pseudomonadota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Bacillus subtilis AQ1
Bacillota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Bacillus sp. PR 1.7
Bacillota |
Bacteria
|
The ability of these arthropods to feed on wood, foliage, and detritus is likely to involve catalysis by different types of cellulases/hemicellulases… |
cellulose hydrolysis
|
|
|
Acinetobacter calcoaceticus strain NRYSBAC-1
Pseudomonadota |
Bacteria
|
Acinetobacter calcoaceticus strain NRYSBAC-1 can degrade Chlorpyrifos and Chlorantraniliprole in vitro. |
pesticide metabolization
|
|
|
Bacillus cereus strain NRYSBBC-1
Bacillota |
Bacteria
|
Bacillus cereus strain NRYSBBC-1 can degrade Chlorpyrifos and Chlorantraniliprole in vitro. |
pesticide metabolization
|
|
|
Klebsiella sp. strain NRYSBKS-1
Pseudomonadota |
Bacteria
|
Klebsiella sp. strain NRYSBKS-1 can degrade Chlorpyrifos and Chlorantraniliprole in vitro. |
pesticide metabolization
|
|
|
Bacillus sp. strain NRYSBBP-1
Bacillota |
Bacteria
|
Bacillus sp. strain NRYSBBP-1 can degrade Chlorpyrifos and Chlorantraniliprole in vitro. |
pesticide metabolization
|
|
|
Bacillus sp. strain NRYSBBS-1
Bacillota |
Bacteria
|
Bacillus sp. strain NRYSBBS-1 can degrade Chlorpyrifos and Chlorantraniliprole in vitro. |
pesticide metabolization
|
Metagenome Information
0 recordsMetagenome sequencing data associated with Scirpophaga incertulas
| Run | Platform | Location | Date | BioProject |
|---|---|---|---|---|
No metagenomes foundNo metagenome records associated with this host species. |
||||
Amplicon Information
12 recordsAmplicon sequencing data associated with Scirpophaga incertulas
| Run | Classification | Platform | Location | Environment |
|---|---|---|---|---|
|
SRR16087858
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087861
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087826
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087828
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087842
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087843
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087814
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087859
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087844
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087825
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087849
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
|
SRR16087860
AMPLICON |
16S
|
-
|
Malaysia
not collected |
-
|
Related Articles
1 recordsResearch articles related to Scirpophaga incertulas
| Title | Authors | Journal | Year | DOI |
|---|---|---|---|---|
|
Goswami, S; Das, SB; Rath, PC ... Mohapatra, SD; Annamalai, M
|
JOURNAL OF ASIA-PACIFIC ENTOMOLOGY
|
2024
|
10.1016/j.aspen.2024.102229 |
Core Microbiome Composition
Core microbiome composition is derived from available metagenomic and amplicon sequencing data, calculated based on the relative abundance and coverage of symbionts across different samples. The representativeness of this analysis may vary depending on the number of available samples and should be considered as a reference guide. See calculation details in Help documentation