Callosobruchus maculatus
cowpea weevil or cowpea seed beetle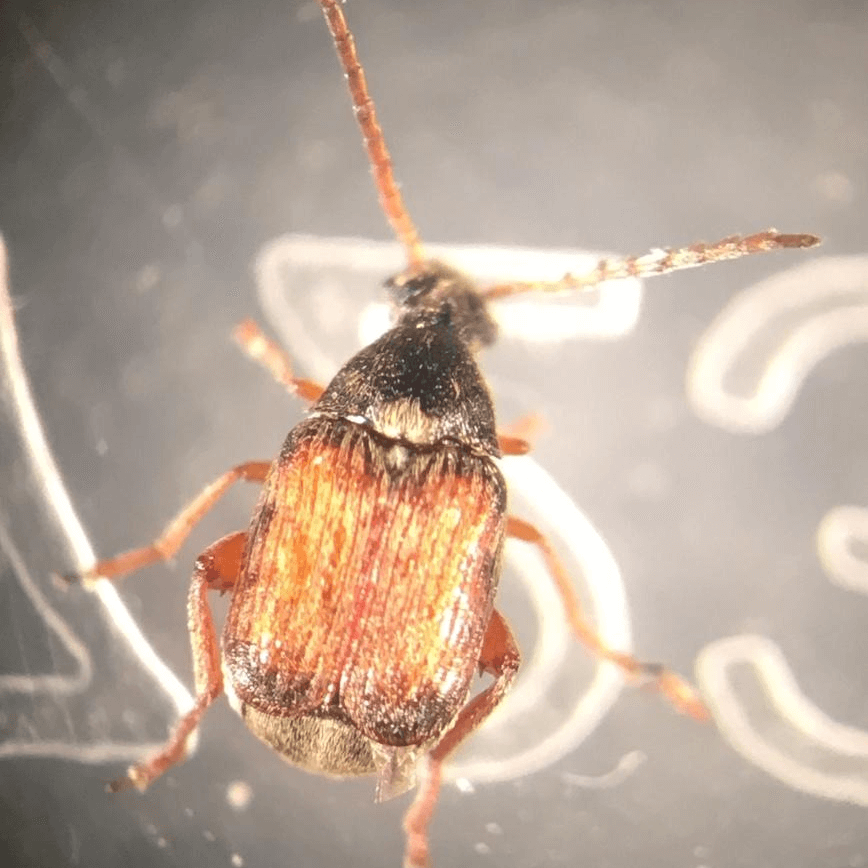
Callosobruchus maculatus is a member of the leaf beetle family, Chrysomelidae, and not a true weevil. This common pest of stored legumes has a cosmopolitan distribution, occurring on every continent except Antarctica.The beetle most likely originated in West Africa and moved around the globe with the trade of legumes and other crops.As only a small number of individuals were likely present in legumes carried by people to distant places, the populations that have invaded various parts of the globe have likely gone through multiple bottlenecks. Despite these bottlenecks and the subsequent rounds of inbreeding, these populations persist. This ability to withstand a high degree of inbreeding has likely contributed to this species’ prevalence as a pest.
Host Genome
Contig| Genome ID | Level | BUSCO Assessment |
|---|---|---|
| GCA_900659725.1 | Contig |
C:87.2%[S:83.2%,D:4.0%],F:4.1%,M:8.7%,n:1367
|
Download Genome Files
Related Symbionts
6 recordsSymbiont records associated with Callosobruchus maculatus
| Classification | Function | Function Tags | Reference | |
|---|---|---|---|---|
|
Staphylococcus gallinarum
Bacillota |
Bacteria
|
Staphylococcus gallinarum encodes complete biosynthetic pathways for the production of B vitamins and amino acids (including tyrosine), and codes for… |
amino acid provision
B vitamin supplementation
carbohydrate metabolism
|
|
|
Bacteria
|
Gut bacteria (These bacterial phyla) may allow adult C. maculatus to survive on DDVP-treated grains, thereby hindering the control of beetle populati… |
pesticide metabolization
|
||
|
Acinetobacter
Pseudomonadota |
Bacteria
|
Acinetobacter may allow adult C. maculatus to survive on DDVP-treated grains, thereby hindering the control of beetle populations in the field. |
pesticide metabolization
|
|
|
Citrobacter
Pseudomonadota |
Bacteria
|
Citrobacter may allow adult C. maculatus to survive on DDVP-treated grains, thereby hindering the control of beetle populations in the field. |
pesticide metabolization
|
|
|
Pseudomonas
Pseudomonadota |
Bacteria
|
Pseudomonas may allow adult C. maculatus to survive on DDVP-treated grains, thereby hindering the control of beetle populations in the field. |
pesticide metabolization
|
|
|
Staphylococcus gallinarum
Bacillota |
Bacteria
|
Staphylococcus gallinarum encodes complete biosynthetic pathways for the production of B vitamins and amino acids (including tyrosine). |
amino acid provision
B vitamin supplementation
|
Metagenome Information
0 recordsMetagenome sequencing data associated with Callosobruchus maculatus
| Run | Platform | Location | Date | BioProject |
|---|---|---|---|---|
No metagenomes foundNo metagenome records associated with this host species. |
||||
Amplicon Information
10 recordsAmplicon sequencing data associated with Callosobruchus maculatus
| Run | Classification | Platform | Location | Environment |
|---|---|---|---|---|
|
SRR13441140
AMPLICON |
16S
|
-
|
Canada
49.81 N 97.13 W |
ENVO:01000219
laboratory insect colonies |
|
SRR8858424
AMPLICON |
16S
|
-
|
Cameroon
7.3276501 N 13.5847197 E |
-
|
|
SRR8858425
AMPLICON |
16S
|
-
|
Cameroon
7.3276501 N 13.5847197 E |
-
|
|
SRR8858426
AMPLICON |
16S
|
-
|
Cameroon
7.3276501 N 13.5847197 E |
-
|
|
SRR8858427
AMPLICON |
16S
|
-
|
Cameroon
7.3276501 N 13.5847197 E |
-
|
|
SRR8858423
AMPLICON |
16S
|
-
|
Cameroon
7.3276501 N 13.5847197 E |
-
|
|
SRR8858429
AMPLICON |
16S
|
-
|
Cameroon
7.3276501 N 13.5847197 E |
-
|
|
SRR8858430
AMPLICON |
16S
|
-
|
Cameroon
7.3276501 N 13.5847197 E |
-
|
|
SRR8858431
AMPLICON |
16S
|
-
|
Cameroon
7.3276501 N 13.5847197 E |
-
|
|
SRR8858428
AMPLICON |
16S
|
-
|
Cameroon
7.3276501 N 13.5847197 E |
-
|
Related Articles
2 recordsResearch articles related to Callosobruchus maculatus
| Title | Authors | Journal | Year | DOI |
|---|---|---|---|---|
|
Berasategui, A; Moller, AG; Weiss, B ... Gerardo, NM; Salem, H
|
APPLIED AND ENVIRONMENTAL MICROBIOLOGY
|
2021
|
10.1128/AEM.00212-21 | |
|
Akami, M; Njintang, NY; Gbaye, OA ... Niu, CY; Nukenine, EN
|
SCIENTIFIC REPORTS
|
2019
|
10.1038/s41598-019-42843-1 |
Core Microbiome Composition
Core microbiome composition is derived from available metagenomic and amplicon sequencing data, calculated based on the relative abundance and coverage of symbionts across different samples. The representativeness of this analysis may vary depending on the number of available samples and should be considered as a reference guide. See calculation details in Help documentation