Acyrthosiphon pisum
pea aphid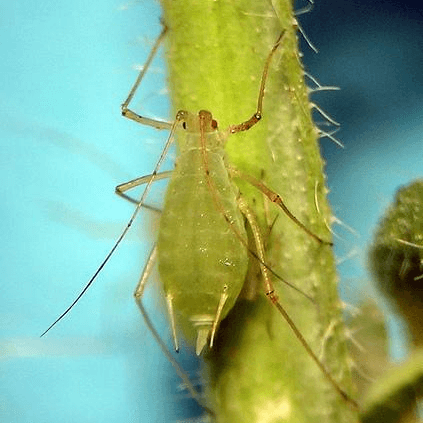
Acyrthosiphon pisum is a sap-sucking insect in the family Aphididae. It feeds on several species of legumes (plant family Fabaceae) worldwide, including forage crops, such as pea, clover, alfalfa, and broad bean, and ranks among the aphid species of major agronomical importance.The pea aphid is a model organism for biological study whose genome has been sequenced and annotated.
Host Genome
Chromosome| Genome ID | Level | BUSCO Assessment |
|---|---|---|
| GCA_005508785.1 | Chromosome |
C:96.2%[S:93.1%,D:3.1%],F:0.8%,M:3.0%,n:1367
|
Download Genome Files
Related Symbionts
57 recordsSymbiont records associated with Acyrthosiphon pisum
| Classification | Function | Function Tags | Reference | |
|---|---|---|---|---|
|
Candidatus Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Candidatus Hamiltonella defensa modifies the anti-predator behavior of Acyrthosiphon pisum by causing the aphids to exhibit proportionately fewer eva… |
natural enemy resistance
|
|
|
Serratia symbiotica
Pseudomonadota |
Bacteria
|
Serratia symbiotica improves the host aphid (Acyrthosiphon pisum) growth and fecundity while reducing longevity. It also provides defense against the… |
growth regulation
fertility
natural enemy resistance
|
|
|
Serratia symbiotica
Pseudomonadota |
Bacteria
|
Serratia symbiotica inhibits the process of regression from winged to wingless morph (apterization) in Acyrthosiphon pisum, increases the body weight… |
growth regulation
developmental modulation
|
|
|
Fukatsuia
Gammaproteobacteria |
Bacteria
|
Fukatsuia symbiotica provides defense against fungal pathogens but interferes with host embryonic development and reproduction, especially at warmer … |
natural enemy resistance
growth regulation
developmental modulation
|
|
|
Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Hamiltonella defensa has a weak effect on the ability of Acyrthosiphon pisum to defend themselves against parasitic wasps Aphidius ervi during the at… |
natural enemy resistance
|
|
|
Regiella insecticola
Pseudomonadota |
Bacteria
|
Regiella insecticola has a weak effect on the ability of Acyrthosiphon pisum to defend themselves against parasitic wasps Aphidius ervi during the at… |
natural enemy resistance
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola (obligate symbiont) is under metabolic constraints or antisense transcripts may reduce its production of pantothenate, resulting … |
vitamin supplementation
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola supplies the host Acyrthosiphon pisum with vitamins and essential amino acids, such as arginine and methionine, that aphids canno… |
detoxification enzymes
|
|
|
Serratia
Pseudomonadota |
Bacteria
|
Serratia provides the host aphid (Acyrthosiphon pisum) with defense against the predator P. japonica by impeding the predator's development and preda… |
natural enemy resistance
|
|
|
Regiella insecticola
Pseudomonadota |
Bacteria
|
Candidatus Regiella insecticola protects pea aphids from the aphid-specific fungal entomopathogen Zoophthora occidentalis but not from the generalist… |
pathogen resistance
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola (the nutritional endosymbiont of Acyrthosiphon pisum) requires aspartate from the host to biosynthesize the essential amino acids… |
amino acid provision
|
|
|
Candidatus Regiella insecticola 5.15
Pseudomonadota |
Bacteria
|
Candidatus Regiella insecticola 5.15 provides strong protection against parasitoid wasps with limited or non-significant negative effects on host sur… |
natural enemy resistance
|
|
|
Serratia symbiotica
Pseudomonadota |
Bacteria
|
The absence of a single naturally occurring isolate of Serratia symbiotica led to a significant preference by wasps for plants that had been fed on b… |
natural enemy resistance
|
|
|
Candidatus Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Candidatus Hamiltonella defensa ameliorates the effects of parasitoid wasp attack by causing the wasp larva to die prematurely, allowing the aphid ho… |
natural enemy resistance
|
|
|
Candidatus Hamiltonella defensa 5AT
Pseudomonadota |
Bacteria
|
Candidatus Hamiltonella defensa 5AT can block the larval development of solitary endoparasitoid wasps (Aphidius ervi and Aphidius eadyi), thereby res… |
natural enemy resistance
|
|
|
Rickettsiella
Pseudomonadota |
Bacteria
|
Rickettsiella changes the insects' body color from red to green by increasing blue-green polycyclic quinones while having less effect on yellow-red c… |
pigmentation alteration
|
|
|
Rickettsiella
Pseudomonadota |
Bacteria
|
Rickettsiella (in Acyrthosiphon pisum) protects against the entomopathogen Pandora neoaphidis by reducing mortality and decreasing fungal sporulation… |
pathogen resistance
|
|
|
Spiroplasma
Mycoplasmatota |
Bacteria
|
Spiroplasma (in Acyrthosiphon pisum) protects against the entomopathogen Pandora neoaphidis by reducing mortality and decreasing fungal sporulation o… |
pathogen resistance
|
|
|
Rickettsia
Pseudomonadota |
Bacteria
|
Rickettsia (in Acyrthosiphon pisum) protects against the entomopathogen Pandora neoaphidis by reducing mortality and decreasing fungal sporulation on… |
pathogen resistance
|
|
|
Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Hamiltonella defensa attenuates the systemic release of volatiles by plants after aphid attack, reducing parasitic wasp recruitment and increasing ap… |
natural enemy resistance
plant defense modulation
|
|
|
Regiella
Pseudomonadota |
Bacteria
|
Regiella (in Acyrthosiphon pisum) protects against the entomopathogen Pandora neoaphidis by reducing mortality and decreasing fungal sporulation on d… |
pathogen resistance
|
|
|
Rickettsiella sp.
Pseudomonadota |
Bacteria
|
The absence of a single-injected isolate of Rickettsiella sp. also led to an attraction of wasps to plants fed on by aphids without secondary symbion… |
natural enemy resistance
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola complements aphid genes in purine metabolism, and the bacterium can meet its nucleotide requirement from aphid-derived guanosine. |
amino acid provision
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola produces essential amino acids (EAAs), matching the supply of bacterial nutrients to the nutritional demand of the animal host. |
amino acid provision
|
|
|
Hamiltonella phage APSE
Pseudomonadota |
Bacteria
|
Hamiltonella phage APSE confers protection against parasitoid wasps, but only when Candidatus Hamiltonella defensa is itself infected by the phage. |
natural enemy resistance
|
|
|
Serratia symbiotica
Pseudomonadota |
Bacteria
|
Serratia symbiotica impairs plant defence response by suppressing Ca2+ elevation and ROS accumulation, allowing colonization of aphids on plants. |
plant defense modulation
|
|
|
Serratia symbiotica
Pseudomonadota |
Bacteria
|
Serratia symbiotica promotes the development and growth of the pea aphid host Acyrthosiphon pisum through enhancing fatty acid biosynthesis. |
growth regulation
developmental modulation
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola (an endosymbiont) can synthesize and provide essential nutrients (e.g., amino acids) for its host, Acyrthosiphon pisum. |
amino acid provision
|
|
|
Candidatus Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Candidatus Hamiltonella defensa substantially affected the hyperparasitoid (either Aphidius ervi or Aphelinus abdominalis) hatch rate. |
natural enemy resistance
|
|
|
Spiroplasma
Mycoplasmatota |
Bacteria
|
The absence of Spiroplasma led to a significant preference by wasps for plants previously attacked by aphids without this symbiont. |
natural enemy resistance
|
|
|
Fukatsuia
Pseudomonadota |
Bacteria
|
Fukatsuia (facultative symbiont) aided the recovery of the obligate symbiont and the host Acyrthosiphon pisum after heat stress. |
temparature adaptation
|
|
|
Regiella
Pseudomonadota |
Bacteria
|
Regiella (facultative symbiont) aided the recovery of the obligate symbiont and the host Acyrthosiphon pisum after heat stress. |
temparature adaptation
|
|
|
Rickettsiella viridis
Pseudomonadota |
Bacteria
|
Rickettsiella viridis infection causes young red aphid larvae to become greener at adulthood, which can reduce predation risk. |
pigmentation alteration
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola (APS) produces arginine, compensating for the pea aphid's lack of capacity to synthesize this amino acid. |
amino acid provision
|
|
|
Serratia symbiotica
Pseudomonadota |
Bacteria
|
Serratia symbiotica infection made aphids more susceptible to most of the tested insecticides than non-infected aphids. |
other
|
|
|
Serratia symbiotica SAp
Pseudomonadota |
Bacteria
|
Serratia symbiotica SAp is closely related to an obligate endosymbiont and is involved in the provision of amino acids. |
amino acid provision
|
|
|
Candidatus Regiella insecticola
Pseudomonadota |
Bacteria
|
Candidatus Regiella insecticola reduces winged offspring production and changes the timing of sexual morph production. |
other
|
|
|
Serratia symbiotica
Pseudomonadota |
Bacteria
|
Serratia symbiotica enzymes may facilitate the digestion of plant proteins, thereby helping to suppress plant defense. |
protein degradation
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola endosymbionts are degraded through a lysosomal-dependent mechanism in senescent bacteriocytes. |
other
|
|
|
Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Candidatus Hamiltonella defensa and Rickettsiella viridis co-infection causes aphid larvae to become darker green. |
pigmentation alteration
|
|
|
Candidatus Regiella insecticola 5.15
Pseudomonadota |
Bacteria
|
Candidatus Regiella insecticola 5.15 significantly reduced parasitoid success and increased aphid survivorship. |
natural enemy resistance
|
|
|
Xenorhabdus bovienii
Pseudomonadota |
Bacteria
|
Xenorhabdus bovienii has the gene PIN1 encoding a protease inhibitor protein active against aphids. |
plant defense modulation
|
|
|
Candidatus Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Candidatus Hamiltonella defensa provides protection against parasitism by the wasp, Aphidius ervi. |
natural enemy resistance
|
|
|
Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Hamiltonella defensa kills parasitoid wasp larvae, allowing aphid hosts to survive and reproduce. |
natural enemy resistance
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola is involved in the biosynthesis of more than 10 essential amino acids. |
amino acid provision
|
|
|
Rickettsiella
Pseudomonadota |
Bacteria
|
Rickettsiella changes the insects' body color from red to green in natural populations. |
pigmentation alteration
|
|
|
Candidatus Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Candidatus Hamiltonella defensa confers protection against parasitoid wasps. |
natural enemy resistance
|
|
|
Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Hamiltonella defensa is the source of resistance to A. ervi parasitism. |
natural enemy resistance
|
|
|
Hamiltonella defensa
Pseudomonadota |
Bacteria
|
Hamiltonella defensa decreased adult survival on Acyrthosiphon pisum. |
growth regulation
|
|
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
Buchnera aphidicola synthesizes the essential amino acid tryptophan. |
amino acid provision
|
|
|
Regiella insecticola
Pseudomonadota |
Bacteria
|
Regiella insecticola protects the aphid from a pathogen. |
other
|
|
|
Buchnera
Pseudomonadota |
Bacteria
|
Buchnera synthesizes amino acids. |
amino acid provision
|
|
|
bacteria
- |
Bacteria
|
- |
||
|
Buchnera aphidicola
Pseudomonadota |
Bacteria
|
- |
||
|
Candidatus Regiella insecticola strain Tut
Pseudomonadota |
Bacteria
|
- |
||
|
Candidatus Serratia symbiotica Strain IS
Pseudomonadota |
Bacteria
|
- |
||
|
Spiroplasma
Mycoplasmatota |
Bacteria
|
- |
Metagenome Information
0 recordsMetagenome sequencing data associated with Acyrthosiphon pisum
| Run | Platform | Location | Date | BioProject |
|---|---|---|---|---|
No metagenomes foundNo metagenome records associated with this host species. |
||||
Amplicon Information
3 recordsAmplicon sequencing data associated with Acyrthosiphon pisum
Related Articles
41 recordsResearch articles related to Acyrthosiphon pisum
| Title | Authors | Journal | Year | DOI |
|---|---|---|---|---|
|
Arai, H; Legeai, F; Kageyama, D; Sugio, A; Simon, JC
|
FEMS MICROBIOLOGY LETTERS
|
2024
|
10.1093/femsle/fnae027 | |
|
Wang, ZW; Zhao, J; Li, GY ... Ye, C; Wang, JJ
|
Insect Science
|
2024
|
10.1111/1744-7917.13315 | |
|
Liu, HP; Yang, QY; Liu, JX ... Liu, CZ; Lv, N
|
FRONTIERS IN PLANT SCIENCE
|
2023
|
10.3389/fpls.2023.1288997 | |
|
Humphreys, RK; Ruxton, GD; Karley, AJ
|
ENTOMOLOGIA EXPERIMENTALIS ET APPLICATA
|
2022
|
10.1111/eea.13223 | |
|
Nozaki, T; Shigenobu, S
|
SCIENTIFIC REPORTS
|
2022
|
10.1038/s41598-022-12836-8 | |
|
Kang, ZW; Zhang, M; Cao, HH ... Liu, FH; Liu, TX
|
MICROBIOLOGY SPECTRUM
|
2022
|
10.1128/spectrum.04066-22 | |
|
Zhou, XF; Ling, XY; Guo, HJ ... Ge, F; Sun, YC
|
INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES
|
2021
|
10.3390/ijms22115951 | |
|
Sochard, C; Bellec, L; Simon, JC; Outreman, Y
|
CURRENT ZOOLOGY
|
2021
|
10.1093/cz/zoaa053 | |
|
Nikoh, N; Tsuchida, T; Koga, R ... Hattori, M; Fukatsu, T
|
Microbiology Resource Announcements
|
2020
|
10.1128/mra.00598-20 | |
|
Wang, QY; Yuan, EL; Ling, XY ... Ge, F; Sun, YC
|
PLANT CELL AND ENVIRONMENT
|
2020
|
10.1111/pce.13836 | |
|
Heyworth, ER; Smee, MR; Ferrari, J
|
FRONTIERS IN ECOLOGY AND EVOLUTION
|
2020
|
10.3389/fevo.2020.00056 | |
|
Blow, F; Bueno, E; Clark, N ... Schmitz, RA; Douglas, AE
|
JOURNAL OF INSECT PHYSIOLOGY
|
2020
|
10.1016/j.jinsphys.2020.104092 | |
|
Skaljac, M; Vogel, H; Wielsch, N; Mihajlovic, S; Vilcinskas, A
|
FRONTIERS IN PHYSIOLOGY
|
2019
|
10.3389/fphys.2019.00438 | |
|
Nikoh, N; Koga, R; Oshima, K; Hattori, M; Fukatsu, T
|
Microbiology Resource Announcements
|
2019
|
10.1128/mra.00272-19 | |
|
Lv, N; Wang, L; Sang, W; Liu, CZ; Qiu, BL
|
Insects
|
2018
|
10.3390/insects9040161 | |
|
Simonet, P; Gaget, K; Balmand, S ... Callaerts, P; Calevro, F
|
Proceedings of the National Academy of Sciences of the United States of America
|
2018
|
10.1073/pnas.1720237115 | |
|
Skaljac, M; Kirfel, P; Grotmann, J; Vilcinskas, A
|
PEST MANAGEMENT SCIENCE
|
2018
|
10.1002/ps.4881 | |
|
Frago, E; Mala, M; Weldegergis, BT ... Gols, R; Dicke, M
|
NATURE COMMUNICATIONS
|
2017
|
10.1038/s41467-017-01935-0 | |
|
McLean, AHC; Hrcek, J; Parker, BJ; Godfray, HCJ
|
ECOLOGICAL ENTOMOLOGY
|
2017
|
10.1111/een.12424 | |
|
Chong, RA; Moran, NA
|
PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA
|
2016
|
10.1073/pnas.1610749113 | |
|
Gauthier, JP; Outreman, Y; Mieuzet, L; Simon, JC
|
PLOS ONE
|
2015
|
10.1371/journal.pone.0120664 | |
|
Polin, S; Le Gallic, JF; Simon, JC; Tsuchida, T; Outreman, Y
|
PLOS ONE
|
2015
|
10.1371/journal.pone.0143728 | |
|
Russell, CW; Poliakov, A; Haribal, M ... van Wijk, KJ; Douglas, AE
|
PROCEEDINGS OF THE ROYAL SOCIETY B-BIOLOGICAL SCIENCES
|
2014
|
10.1098/rspb.2014.1163 | |
|
Lukasik, P; van Asch, M; Guo, HF; Ferrari, J; Godfray, HCJ
|
ECOLOGY LETTERS
|
2013
|
10.1111/ele.12031 | |
|
Parker, BJ; Spragg, CJ; Altincicek, B; Gerardo, NM
|
APPLIED AND ENVIRONMENTAL MICROBIOLOGY
|
2013
|
10.1128/AEM.03193-12 | |
|
Weldon, SR; Strand, MR; Oliver, KM
|
PROCEEDINGS OF THE ROYAL SOCIETY B-BIOLOGICAL SCIENCES
|
2013
|
10.1098/rspb.2012.2103 | |
|
Zeng, FR; Xue, RF; Zhang, HQ; Jiang, TZ
|
PEST MANAGEMENT SCIENCE
|
2012
|
10.1002/ps.3299 | |
|
Manzano-Marín, A; Lamelas, A; Moya, A; Latorre, A
|
PLOS ONE
|
2012
|
10.1371/journal.pone.0047274 | |
|
Hansen, AK; Vorburger, C; Moran, NA
|
GENOME RESEARCH
|
2012
|
10.1101/gr.125351.111 | |
|
Hansen, AK; Moran, NA
|
Proceedings of the National Academy of Sciences of the United States of America
|
2011
|
10.1073/pnas.1013465108 | |
|
Ramsey, JS; MacDonald, SJ; Jander, G ... Thomas, GH; Douglas, AE
|
INSECT MOLECULAR BIOLOGY
|
2010
|
10.1111/j.1365-2583.2009.00945.x | |
|
Wilson, ACC; Ashton, PD; Calevro, F ... Thomas, GH; Douglas, AE
|
INSECT MOLECULAR BIOLOGY
|
2010
|
10.1111/j.1365-2583.2009.00942.x | |
|
Tsuchida, T; Koga, R; Horikawa, M ... Simon, JC; Fukatsu, T
|
SCIENCE
|
2010
|
10.1126/science.1195463 | |
|
Degnan, PH; Yu, Y; Sisneros, N; Wing, RA; Moran, NA
|
PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA
|
2009
|
10.1073/pnas.0900194106 | |
|
Degnan, PH; Moran, NA
|
MOLECULAR ECOLOGY
|
2008
|
10.1111/j.1365-294X.2007.03616.x | |
|
Oliver, KM; Campos, J; Moran, NA; Hunter, MS
|
PROCEEDINGS OF THE ROYAL SOCIETY B-BIOLOGICAL SCIENCES
|
2008
|
10.1098/rspb.2007.1192 | |
|
Leonardo, TE; Mondor, EB
|
PROCEEDINGS OF THE ROYAL SOCIETY B-BIOLOGICAL SCIENCES
|
2006
|
10.1098/rspb.2005.3408 | |
|
Moran, NA; Degnan, PH; Santos, SR; Dunbar, HE; Ochman, H
|
PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA
|
2005
|
10.1073/pnas.0507029102 | |
|
Scarborough, Claire L.; Ferrari, Julia; Godfray, H. C. J.
|
Science
|
2005
|
10.1126/science.1120180 | |
|
Oliver, KM; Moran, NA; Hunter, MS
|
Proceedings of the National Academy of Sciences of the United States of America
|
2005
|
10.1073/pnas.0506131102 | |
|
LAI, CY; BAUMANN, L; BAUMANN, P
|
Proceedings of the National Academy of Sciences
|
1994
|
10.1073/pnas.91.9.3819 |
Core Microbiome Composition
Core microbiome composition is derived from available metagenomic and amplicon sequencing data, calculated based on the relative abundance and coverage of symbionts across different samples. The representativeness of this analysis may vary depending on the number of available samples and should be considered as a reference guide. See calculation details in Help documentation